PAOC Spotlights
Big Oceans. Tiny Microbes. Sea Biomes.
The Simons Foundation extends its support for microbial oceanography with the establishment of the Simons Foundation Collaboration on Ocean Computational Biogeochemical.
Microbes sustain all of Earth’s habitats, including its largest biome, the global ocean. Microbes in the sea capture solar energy, catalyze biogeochemical transformations of important elements, produce and consume greenhouse gases, and fuel the marine food web. Measuring and modeling the distribution, composition and function of microbial communities, and their interactions with the environment, are key to understanding these fundamental processes in the ocean.
The Simons Foundation, which provides generous funding for several lines of research within EAPS, recently extended its support for microbial oceanography with the establishment of the Simons Foundation Collaboration on Ocean Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES.) Led by MIT Professor of Oceanography Michael Follows, CBIOMES draws together an multidisciplinary group of both U.S. and international investigators bridging oceanography, statistics, data science, ecology, biogeochemistry, and remote sensing.
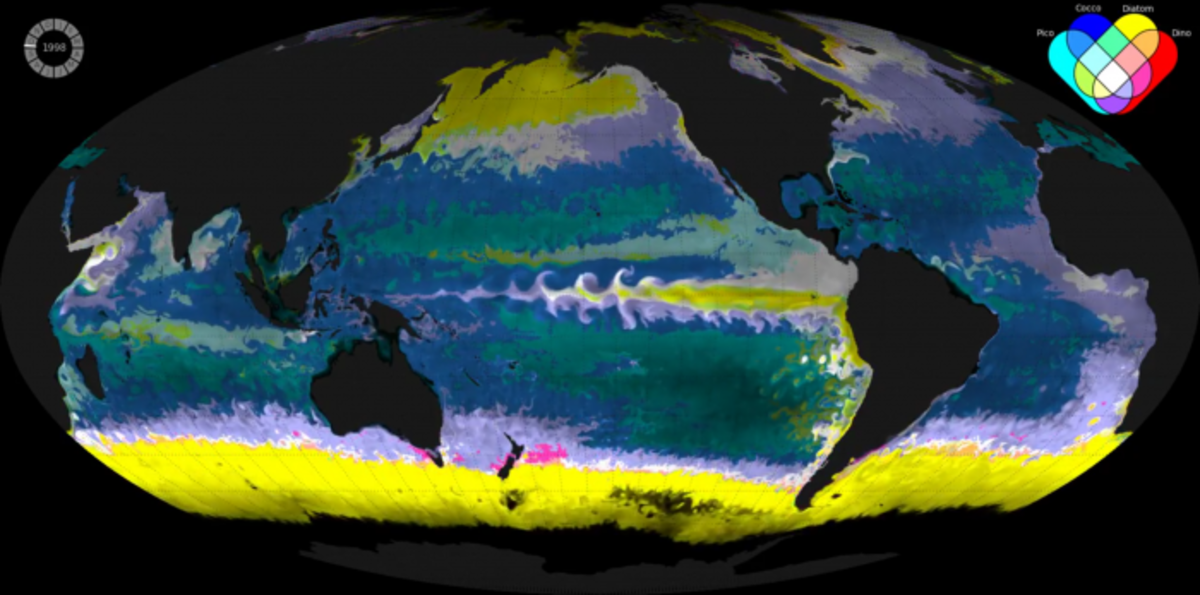
Ecological Provinces from a computer simulation of the marine global ecosystems developed in the Follows Group at MIT. Each color represents a different distinct combination of the most dominant phytoplankton function types (as shown in the Venn diagram top right). The opacity indicates the total concentration of phytoplankton biomass: the darker the color, the less phytoplankton. (Credit: Oliver Jahn, MIT)
The goal of CBIOMES (pronounced “sea biomes”), which leverages and extends Follow’s Darwin Project activity, is to develop and apply quantitative models of the structure and function of marine microbial communities at seasonal and basin scales.
As Follows explains, “Microbial communities in the sea mediate the global cycles of elements including climatically significant carbon, sulfur and nitrogen. Photosynthetic microbes in the surface ocean fix these elements into organic molecules, fueling food webs that sustain fisheries and most other life in the ocean. Sinking and subducted organic matter is remineralized and respired in the dark, sub-surface ocean, maintaining a store of carbon about three times the size of the atmospheric inventory of CO2.”
The communities of microbes that sustain these global-scale cycles are functionally and genetically extremely diverse, non-uniformly distributed and sparsely sampled. Their biogeography reflects selection according to the relative fitness of myriad combinations of traits that govern interactions with the environment and other organisms. Trait-based theory and simulations provide tools with which to interpret biogeography and microbial mediation of biogeochemical cycles. Follows says, “Several outstanding challenges remain: Observations to constrain the biogeography of marine microbes are still sparse and based on eclectic sampling methods. Theories of the organization of the system have not been quantitatively tested, and the models used to simulate the system still lack sufficiently mechanistic biological foundations. Addressing these issues will enable meaningful, dynamic simulations and state estimation.”
CBIOMES seeks to integrate key new data sets in real time as they are collected at sea to facilitate direct tests of theoretical predictions to synthesize an atlas of marine microbial biogeography suitable for testing a range of specific ecological theories and quantifying the skill of numerical simulations. It also aspires to develop new trait-based models and simulations of regional and global microbial communities bringing to bear the power of metabolic constraints and knowledge of macro-molecular composition; to analyze these data and models using statistical tools to interpolate and extrapolate the sparse data sets, formally quantify the skill of numerical simulations, and employ data assimilation technologies to identify and optimize compatible model frameworks. “Together, the results of these efforts will advance new theoretical approaches and lead to improved global ocean-scale predictions and regional state-estimates, constrained by observed biogeography. They will provide a quantification of the associated biogeochemical fluxes,” says Follows.
Working with Follows on CBIOMES are PIs Stephanie Dutkiewicz (MIT), Jacob Bien, Christopher Edwards, and Jed Fuhrman (USC), Zoe Finkel and Andrew Irwin (Mt Allison University, New Brunswick), Shubha Sathyendranath(Plymouth Marine Laboratory, UK), and Joseph Vallino (MBL, Woods Hole.)
A meeting held at the Simons Foundation in New York City May 21 through 23 provided a first opportunity for collaborators to meet face-to-face, and provided a forum for investigators to educate one another about each others expertise and areas of activity, share initial progress, and coordinate collaborative efforts.
Discussions centered around how to determine the biogeography of marine microbes from empirical date, the role of statistical models in determining the relationships in space and time between organisms, traits, and environments, the complimentary role of mechanistic models and how to simulate the systems that are observed, and, in the context of model-date synthesis, how to best utilize empirical data to test theory and improve simulation skill.
“While the central question `What is the functional biogeography of a group of organisms in the oceans?’ is relatively focused, the techniques being used are extremely varied focusing a lot on computational tools, but uniquely, hand in hand with data collection and data compilation,” says Follows. “I am particularly excited by everyone’s enthusiasm, the number of cross-connections and collaborations already underway, and the rapid progress that is happening on many fronts.”
Complementary to CBIOMES is the Simons Collaboration on Ocean Processes and Ecology (SCOPE) co-led by Ed DeLong and David Karl. SCOPE’s focus is advancing understanding of marine biology, biogeochemistry, ecology and evolution of microbial processes by focusing on a representative ocean benchmark (at Station ALOHA) located in the North Pacific Subtropical Gyre.
SCOPE-Gradients, a related project, with a focus on understanding transitions between the North Pacific Subtropical Gyre and neighboring ecosystems brings a rich stream of observational data to the CBIOMES effort. The North Pacific Subpolar Gyre is a region of open ocean notable for exhibiting steep changes in environmental conditions (gradients) associated with dramatic changes in the microbial ecosystem. Several members of the SCOPE-Gradients team accompanied project PI Virginia Armbrust (UW) to the May CBIOMES meeting where they shared a preliminary implementation of the dynamic atlas they are constructing to curate disparate observational data and model results within a common framework.
About the Simons Foundation
The mission of the Simons Foundation mission is to advance the frontiers of research in mathematics and the basic sciences. Co-founded in New York City by Jim and Marilyn Simons, the foundation exists to support basic — or discovery-driven — scientific research undertaken in the pursuit of understanding the phenomena of our world.
As well as Michael Follows, other Simons Foundation funded investigators in EAPS include Tanja Bosak, Gregory Fournier, and Roger Summons. Several postdoctoral researchers have been recipients of Simons Postdoctoral Fellowships, among them Alexandria Johnson, Sukrit Ranjan and Christopher Follett.
Related