PAOC Spotlights
A systems level approach to biogeography
Read this at CBIOMES
The most abundant photosynthetic organisms on Earth belong to the marine cyanobacterial genus Prochlorococcus. A wide diversity of Prochlorococcus strains can be found at multiple scales, from a single droplet of seawater to ocean basins, presenting a challenge to understanding the links between cellular-scale processes and ecosystem-scale patterns. A new paper drawing together CBIOMES investigators John Casey (lead author), Zoe Finkel, Christian Mueller, Mick Follows, and others, provides a ground-breaking, first of its kind study, demonstrating the potential to interpret and relate global scale ecosystem organization among different strains of Prochlorococcus in terms of cellular and genome-scale processes. The paper appears in Science Advances.
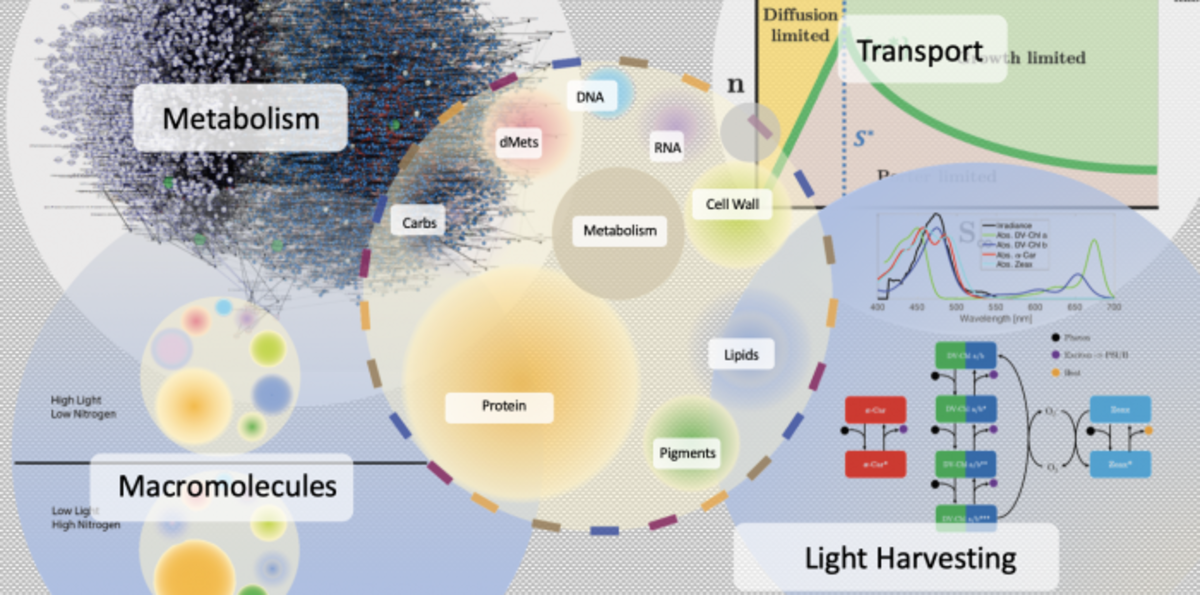
Building from a database of several hundred Prochlorococcus genomes and decades of physiological studies, Casey and co-authors developed a mechanistic modeling framework that simulates the growth, metabolic fluxes, and physiological acclimation processes of all known strains across the environmental state space. The framework, called the Microbial Simulation Environment or MSE (https://www.github.com/jrcasey/mse) was applied to interpret the physiological, metabolic, and genetic underpinnings of Prochlorococcus biogeography along a meridional transect of the North and South Atlantic Oceans (Atlantic Meridional Transect) where prior observations of the environmental context and biogeography of Prochlorococcus ecotypes provide drivers and contextual data.
Emerging from their solutions the team found a remarkable correspondence between predicted patterns of maximal growth rate and observed population densities, confirming, in a real-world context, the often-assumed relationship between growth rate and fitness.
“Despite its global dominance and significance to biogeochemical cycles, we are only just learning what controls the observed global distribution of Prochlorococcus,” says Casey. “Because of the outsized role they play in global marine ecosystems, determining what sets the biogeography of this species is going to be particularly key when we come to think about the prediction of future change. There is a mountain of physiological and genomic knowledge about Prochlorococcus, but it’s when we consolidate everything in a mechanistic, systems-level framework that we can make more interpretable predictions and learn something new.”
The authors are excited that the emergent relationship they find in modeled specific growth rate and observed biomass in their work could provide a calibration for a density-dependent mortality closure with broader application. “A recent study led by Chris Follett and other CBIOMES researchers picked up on a mechanism for top-down control of Prochlorococcus populations at the high latitude frontiers of their distribution; we’re seeing that same phenomenon but we’re detecting it as something that’s missing from a purely bottom-up perspective, so they’re really complementary angles of approach.”
Casey says, “Implementation of this framework in regions where appropriate environmental data are already available can provide testable predictions on the abundance, physiology, and metabolism of Prochlorococcus. Allowing the virtual organisms to feedback on their resource environment could lead to global-scale prognostic applications, perhaps embedded in a coarser-grained general population.” Though currently still computationally prohibitive, the team is confident that with effort on code development and optimization, including such feedback mechanisms is a plausible goal towards exploring microbial community interactions more deeply.
The paper received support through the Simons Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES) which seeks to develop and apply quantitative models of the structure and function of marine microbial communities at seasonal and basin scales.
Story image: Metabolic and physiological acclimation processes simulated by MSE. A single cell (center) is represented by its unique metabolic network (top left), nutrient transporters (top right), macromolecular pools (bottom left), and photosynthetic apparatus (bottom right). MSE allows each cell type to acclimate to a new environment by adjusting these components, subject to strain-specific constraints – adapted by H. Hill.